Using PCR primers with standard Recombinase Polymerase Amplification reagents
Here at TwistDx™ we are constantly working to improve RPA even further, and are discovering new capabilities and features of the reaction. We can now report that in some instances RPA will work straight out of the box with established oligonucleotide PCR primers that are as short as 18 residues, and which require no additional optimisation. To date, we have found that PCR primers will work for amplicons of up to 500 base pairs in length, although incubation times do vary with primer length and amplicon size for gel-based detection.
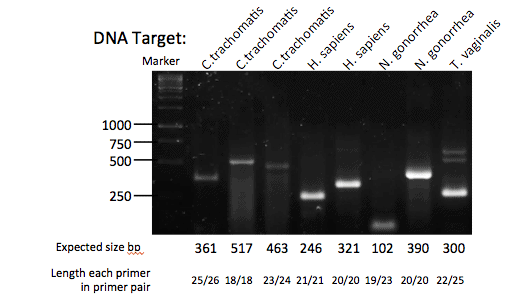
We do still recommend that RPA users design longer primers of 30-35 nucleotides in length to be sure of an optimum reaction, but customers can also try proven PCR primers in an RPA against specific target sequences. It would be great to hear of new instances where using PCR primers work well - contact techsupport@twistdx.co.uk with your feedback.
Below are some examples of RPA reactions that have been established using published PCR primers and 103 copies of target (mainly genomic DNA).
Primer sequences
C. t1 TCTTTTTAAACCTCCGGAACCCACTT/GGATGGCATCGCATAGCATTCTTTG
C. t2 GGACAAATCGTATCTCGG/GAAACCAACTCTACGCTG
C. t3 TGATGCTAGGGACGGATTAAAACC/TTCCCCTAAATTATGCGGTGGAA
H. s1 CGTGTACTTGATCGTCGCCAT/CAAGGAGGAAGGCTAGGGCTA
H. s2 GTGCTGTTTGCAGCTCTGAG/AGTGGCCAAGGAGTCGTATG
N. g1 CCGGAACTGGTTTCATCTGATT/GTTTCAGCGGCAGCATTCA
N. g2 GCTACGCATACCCGCGTTGC/CGAAGACCTTCGAGCAGACA
T. v ATTGTCGAACATTGGTCTTACCCTC/TCTGTGCCGTCTTCAAGTATGC
References:
C. t1 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3854900/
C. t2 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC269699/
C. t3 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2944303/
H. s1 http://pga.mgh.harvard.edu/primerbank/[SK2]
H. s2 http://www.ncbi.nlm.nih.gov/pubmed/12209956
N. g1 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1876173/
N. g2 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC495310/
T. v http://www.ncbi.nlm.nih.gov/pmc/articles/PMC87441/